
Calculating a global alignment is a form of global optimization that "forces" the alignment to span the entire length of all query sequences. Computational approaches to sequence alignment generally fall into two categories: global alignments and local alignments. Instead, human knowledge is primarily applied in constructing algorithms to produce high-quality sequence alignments, and occasionally in adjusting the final results to reflect patterns that are difficult to represent algorithmically (especially in the case of nucleotide sequences). Very short or very similar sequences can be aligned by hand however, most interesting problems require the alignment of lengthy, highly variable or extremely numerous sequences that cannot be aligned solely by human effort. In business, more specifically in marketing, sequences of purchases are also increasingly being analyzed by the same methods as in bioinformatics.
#Compare two amino acid sequences series#
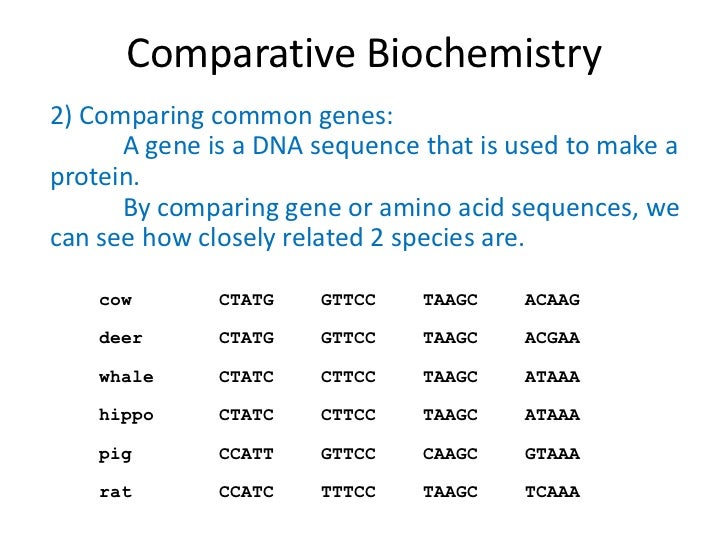
Sequence alignment can be used for non-biological sequences, such as identifying similarities in a series of letters and words present in human language. Although DNA and RNA nucleotide bases are more similar to each other than to amino acids, the conservation of base pairing can indicate a similar functional or structural role. The absence of substitutions, or the presence of only very conservative substitutions (that is, the substitution of amino acids whose side chains have similar biochemical properties) in a particular region of the sequence, suggest that this region has structural or functional importance. In protein sequence alignment, the degree of similarity between amino acids occupying a particular position in the sequence can be interpreted as a rough measure of how conserved a particular region or sequence motif is among lineages. If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another.
A sequence alignment, produced by ClustalW between two human zinc finger proteins identified by GenBank accession number.


 0 kommentar(er)
0 kommentar(er)
